Translate this page into:
Pathotype and virulence of Escherichia coli from adult human diarrheal feces and water sources in Lagos, Nigeria
*Corresponding author: Nwamaka Herrienta Igbokwe, PhD Department of Pharmaceutical Microbiology and Biotechnology, Faculty of Pharmacy, University of Lagos, Lagos, Nigeria. nigbokwe@unilag.edu.ng
-
Received: ,
Accepted: ,
How to cite this article: Igbokwe NH, Idowu AO, Ezeobiora CE, et al. Pathotype and virulence of Escherichia coli from adult human diarrheal feces and water sources in Lagos, Nigeria. Am J Pharmacother Pharm Sci. 2025:003.
Abstract
Objectives:
The prevalence of Escherichia coli strains from fecal diarrheal specimens and water samples underscores the imperative of exploring the symbiotic interplay between microbial ecologies in elucidating the pathophysiology of diarrheal infections among the adult populace. This study determined the different pathotypes of E. coli strains in water and stool samples from a Nigerian state, Lagos, and the different virulence factors they exhibit.
Materials and Methods:
E. coli strains were isolated and characterized by presumptive coliform tests from stool samples of patients with diarrheal and from different water sources in Lagos. Multiplex conventional and real-time polymerase chain reaction (PCR) studies that utilized a comprehensive set of 22 primers that enabled the selective amplification of 11 virulence genes, namely: stx1, stx2, eae, bfp, lt, st11, virF, ipaH, aafII, daaE, and uidA were used to detect different categories of diarrheagenic E. coli pathotypes.
Results:
The real-time and conventional PCR analysis of the 204 E. coli strain from the diarrheal stool and water samples detected the six diarrheagenic E. coli pathotypes (enterohemorrhagic, enteropathogenic, enterotoxigenic, enteroinvasive, enteroaggregative, and diffuse adherent) and their virulence toxins; stx2, stx1, eae, bfp, st11, lt, virF, aafII, daaE, and uidA with an exception of ipaH toxin, the gene for enteroinvasive E. coli, which was not detected. Enterohemorrhagic/enteropathogenic E. coli toxin eae, 18 (32.29%), was the most detected toxin next to uidA which was isolated from all the samples of E. coli strains from Lagos.
Conclusion:
The expression of these virulence genes shows that these organisms exhibit a high degree of pathogenicity, thereby presenting a substantial danger to public health.
Keywords
Escherichia coli
Pathotype
Virulence
Enteropathogenic
Diarrheal
INTRODUCTION
The prevalence of diarrheal illness poses a substantial public health concern.[1-3] especially in developing countries[4-6] where the disease burden is more on children[7-9] and holds precedence as the leading cause of mortality[9-11] with an attributed toll of approximately 2 million morbidities each year worldwide.[12-14] There is a compelling link between diarrheal diseases and water contamination on a global scale.[15,16] The etiologic factors associated with this disease include microbial agents which are usually transmitted through food and water contaminated with human feces.[15,17] Diarrheal disease alone causes 2.2 million of the 3.4 million water-related deaths per year.[18] The features of acute diarrhea vary from place to place depending on geography and socioeconomic variables.[18,19] The major causes of this illness include poor quality of water, poor hand and food hygiene, and poor sanitation[18,19] In developed countries, the mortality rates have declined considerably in recent times due to improvements in general health hygiene and advances in health care. The morbidity persists in so many other countries where outbreaks of diarrheal diseases continue to affect millions of infants, young children, the immune-compromised, and adults.[2] Foreigners visiting tropical developing countries frequently experience traveler’s diarrhea, caused by agents that are endemic in those countries because visitors have not had the opportunity to develop protective immunity against it.[20,21]
Most Escherichia coli strains are commensals,[22] however, some strains have developed the ability to cause diseases of the gastrointestinal tract, urinary tract, or central nervous systems even in the most robust human hosts. Strains that cause enteric infections are designated diarrheagenic E. coli, a group that includes emergent pathogens with public health relevance worldwide. Diarrheagenic E. coli strains can be divided into six main categories based on their distinct molecular, clinical, and pathological features.[23,24] The strains include emergent pathotypes that differ in their virulence factors (toxins). They are enterohemorrhagic E. coli (EHEC), a Shiga toxin-producing E. coli that is characterized by the production of two potent Shiga-like cytotoxins 1 and 2 (Stx1 and Stx2).[25] Infection with EHEC is a leading cause of bloody diarrhea and hemorrhagic colitis, occasionally resulting in life-threatening systemic complications including hemolytic uremic syndrome. Enteropathogenic E. coli (EPEC) harbors a pathogenicity island that encodes a series of proteins involved in the adhering to and expunging alterations of the intestinal microvilli of the host cell.[26] Attachment to the intestinal microvilli is mediated by an outer membrane protein called intimin, encoded by eae, and another adherence gene factor encoded as bundle-forming pili, bfp.[23] Enterotoxigenic E. coli produces heat-labile (LT) or heat-stable (ST) enterotoxins which are generally represented as lt and st11 genes. Enteroinvasive E. coli (EIEC) has biochemical, physiological, and genetic properties similar to those of Shigella. It causes an epithelial invasion of the large bowel leading to inflammation and ulceration of the mucosa.[27] The genes (ipaH, ipaBCD, ial, virF, virB, sigA, set1A, sepA, sat, pic, set1B, and sen) related to invasion are located in a virulence plasmid.[28] Enteroaggregative E. coli (EAggEC) are a heterogeneous group and have been traditionally defined by their characteristic “stacked brick”-like aggregative adherence (AA) to HEp-2 cells.[29-31] This E. coli strain adheres to the intestinal epithelial cells causing biofilm formation, mucus secretion, mucosal inflammation, and cytotoxic damage.[32] The virulence factors involved include the AA fimbriae (AAF) which is coded as aafII gene. Diffuse adherent E. coli (DAEC) are characterized by the presence of Afa/Dr. adhesion, daaE genes.[33] The different E. coli pathotypes exhibit different virulence factors that enhance the disease burden. This study aimed at using the multiplex conventional and real-time polymerase chain reaction (PCR) to determine the virulence toxins and pathotypes of E. coli strains from Lagos, Nigeria.
MATERIALS AND METHODS
Ethical statement
The protocol for the study was reviewed and approved by the Ethics and Human Subjects Committee of the College of Medicine, University of Lagos, Nigeria, with Protocol number: CM/COM/08/VOL.XXV. Study participants and (or) their guardians provided written informed and/or assent and the research was conducted in compliance with the Helsinki Declaration on the conduct of human health research.
Collection of diarrheal stool samples
Adult diarrheal stool samples were collected from seven densely populated localities (Alimosho, Shomolu, Oshodi, Surulere, Lagos Mainland, Ikeja, and Mushin) in Lagos state as indicated in Figure 1. Stool samples were collected from patients suffering from Diarrhea (age range, 18–65 years) who reported at the outpatient units of general hospitals in each of the sampled localities in Lagos and some medical laboratories in the localities between April 2011 and March 2012. Fresh fecal samples were immediately placed in a specialized transport medium (Stuart’s transport medium, Oxoid®, Basingstoke, United Kingdom) to maintain the freshness and the viability of microorganisms during transit. The samples were kept on ice while being transported to the laboratory for subsequent same-day analysis.
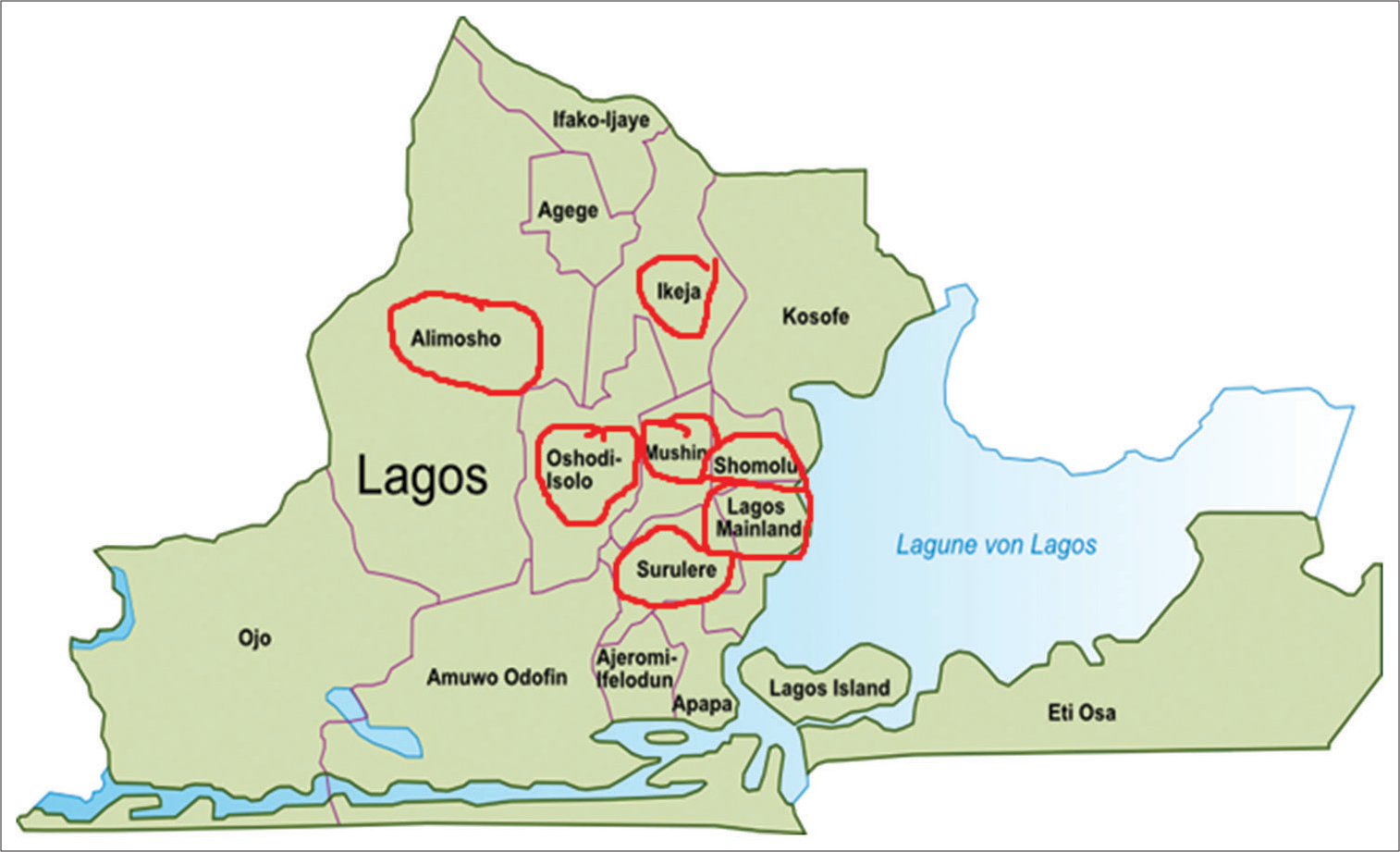
- Map of Lagos state indicating the localities (circled in red) from where stool and water samples were collected.
Control stool specimens were obtained from apparently healthy adults while informed consent of the patients used in this study was obtained. None of the patients reported receiving antibiotic therapy <3 weeks before sampling. Presumptive coliform tests for isolation and characterization of E. coli strains from diarrheal stool samples and water samples were carried out in the Department of Pharmaceutical Microbiology Laboratory, Faculty of Pharmacy, University of Lagos and the Department of Pharmaceutical Microbiology Laboratory, Faculty of Pharmacy, Obafemi Awolowo University, Ile-Ife, Osun state, Nigeria. The protocol involved the isolation of one E. coli strain per sample.
Adult diarrheal stool samples and control (from volunteered healthy individuals), water samples, and the isolated E. coli strains from Lagos were transported on ice in March 2012 to the city of Milwaukee Health Department, United States of America for further investigations.
Collection of water samples
Sachet “pure water” collection
Commercial sachets “pure water produced in the locality under study were purchased from the open markets in each of the stated localities in Lagos.”
Tap water collection
The tap faucets were flamed, turned on, and allowed to flow for 5 min. 100 mL of water was collected in a sterile bottle container; the bottleneck was flamed, covered, and sent to the laboratory for analysis.
Well water
The sterile bottle was dipped into the well, stirred, and pulled out carefully, avoiding the well edges when filled with water.
Canal water collection
The samples collected from open drains and canals must be representative of the areas being sampled. This was therefore achieved by collecting, pooling together, and stirring water samples from various locations and depths in the canal. The sterile screwed sample bottles were dipped into various locations of the canals, swirled, and filled with water for analysis.
Isolation of E. coli from diarrheal fecal samples and aqueous samples
The combined methods of the conventional Clinical and Laboratory Standards Institute and Peter et al.[34,35] were used in this investigation which involved culturing and subculturing into various diagnostic media for enteric bacterial pathogens for each of the samples (diarrheal stool and water). The samples were introduced into peptone water (Oxoid®) and then placed in an incubator set at 37°C for 24 h. Swabs of stool samples were swirled in 5 mL MacConkey broth containing Durham tubes and incubated for 24 h at 37°C. From each of the broths showing acid and gas, subcultures were made into clearly labeled MacConkey agar, blood agar, and eosin methylene blue agar (EMBA) (Oxoid, Basingstoke, England) and placed in an incubator for 24 h at 37°C. One colony from either of the plates with distinct E. coli morphologies was subcultured in freshly prepared EMBA and incubated for 24 h at 37°C. For the water samples, 1 mL of 1:106 dilution of the canal water, 1 mL of well, dechlorinated tap, and sachet water each was subcultured in 5 mL MacConkey broth and incubated for 24 h. After 24 h of incubation, swabs of each sample were streaked on MacConkey agar and incubated for 24 h. A colony from each of the samples showing clear morphology of E. coli was streaked on separate clearly labeled EMBA plates and incubated for 24 h. From EMBA plates, a colony of each strain was transferred into blood agar and incubated for 18 h. The isolated E. coli strains were typed using the analytical profile index (API)[36] which involved the use of 20 E strips (Bio Merieux®, Marcy-I’Etiole, France). The characterized E. coli strains were stored in duplicate motility agar and 20%v/v glycerol and trypticase soy broth (TSB).
Characterization and typing of E. coli strains using the API
API 20 E V 4.0 test kit (Biomerieux, USA) for Enterobacteriaceae was used for the analysis. A total of 2–5 colonies of 18-h blood agar culture of each suspected E. coli strain were suspended in 5 mL of normal saline. Incubation boxes (tray and lid) were prepared by distributing 5 mL of distilled water into the honeycombed wells of the tray to create a humid atmosphere and strips were placed in each of them. The identification of each isolate was recorded on the elongated flap of the designated tray. The bacterial suspension of each strain was carefully distributed into the tubes of their corresponding strips according to the manufacturer’s instructions. The closed incubation boxes were incubated for 18 h at 37°C. After incubation, specific reagents were added to some of the tubes and color reactions were observed as positive or negative after 10 min. Identification was obtained using the API identification software and also referring to the manual table. The confirmed E. coli strains were stored in duplicates in (i) motility agar and (ii) 20%v/v glycerol + TSB.
Nucleic acid extraction from E. coli strains
DNA from E. coli was extracted from whole organisms by boiling using the previously described method[23] with slight modification. E. coli strains were sub-cultured onto blood agar and incubated for 18 h. One colony of each strain was suspended in 500 µL of PCR-grade water in a capped tube and boiled at 100°C for 10 min. The tubes were removed from boiling and placed directly on ice for 2 min. The lysate was centrifuged at 4°C, 14000 rpm for 10 min. 400 µL of the supernatant was stored at −80°C for further investigations.
The methodology outlined by Vidal et al.[23] was utilized for the amplification and identification of virulence toxin genes through conventional multiplex real-time PCR.
Single multiplex PCR detection of E. coli virulence toxins
The elaboration and detection of the genes encoding toxins and factors contributing to pathogenicity were done by real-time PCR using a conventional multiplexing technique previously described.[23] Diarrheagenic E. coli target toxins include enterohemorrhagic, Shiga toxin-producing (stx1, stx2) genes, enteropathogenic (eae, bfp) genes, enterotoxigenic (stII and lt) genes, enteroinvasive (virF and ipaH) genes, enteroaggregative (aafII) gene, and diffuse adherent (daaE) gene. This technique utilized a comprehensive set of 22 primers that enabled the selective amplification of 11 virulence genes as shown in Table 1. Diarrheagenic E. coli reference strains, 011 (stx1 and stx2), 2348/69 (bfp and eae), H10407 (st and lt), EI-34 Nal-R (ipaH and virF), EI 34 Strep-R (ipaH and virF), O42 (aafII), 933J (stx1, stx11, eae), and F-1845 (daaE), were used as positive controls while Shigella flexneri and PCR grade water were used as negative controls [Figure 1].
| Strain | Toxin | Primers | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| UidAF | 5’d CAGTCTGGATCGCGAAAACTG 3’ | 180 | [37] | |
| UidAR | 5’d ACCAGACGTTGCCCACATAATT 3’ | |||
| EHEC | St×1F | 5’d CAGTTAATGTGGTGGCGAAGG 3’ | 348 | [9] |
| St×1R | 5’d CACCAGACCATGTAACCGCTG 3’ | |||
| EHEC | St×2F | 5’d ATCCTATTCCCGGGAGTTTACG 3’ | 582 | [9] |
| St×2R | 5’d GCGTCATCGTATACACAGGAGC 3’ | |||
| EPEC | EaeF | 5’d TCAATGCAGTTCCGTTATCAGTT 3’ | 482 | [38] |
| EaeR | 5’d GTAAAGTCCGTTACCCCAACCTG 3’ | |||
| EPEC | BfpF | 5’d GGAAGTCAAATTCATGGGGGTAT 3’ | 300 | [9] |
| BfpR | 5’d GGAATCAGACGCAGACTGGTAGT 3’ | |||
| ETEC | LTF | 5’d GCACACGGAGCTCCTCAGTC 3’ | 218 | [39] |
| LTR | 5’d TCCTTCATCCTTTCAATGGCTTT 3’ | |||
| ETEC | STIIF | 5’d AAAGGAGAGCTTCGTCACATTTT 3’ | 129 | [9] |
| STIIR | 5’d AATGTCCGTCTTGCGTTAGGAC 3’ | |||
| EIEC | VirF | 5’d AGCTCAGGCAATGAAACTTTGAC 3’ | 618 | [23] |
| VirR | 5’d TGGGCTTGATATTCCGATAAGTC 3’ | |||
| EIEC | IpaHF | 5’d CTCGGCACGTTTTAATAGTCTGG 3’ | 933 | [23] |
| IpaHR | 5’d GTGGAGAGCTGAAGTTTCTCTGC 3’ | |||
| DAEC | DaaEF | 5’d GAACGTTGGTTAATGTGGGGTAA 3’ | 542 | [23] |
| DaaER | 5’d TATTCACCGGTCGGTTATCAGT 3’ | |||
| EAggEC | AafIIF | 5’d CACAGGCAACTGAAATAAGTCTGG 3’ | 378 | [23] |
| AafIIR | 5’d ATTCCCATGATGTCAAGCACTTC 3’ |
F: Forward, R: Reverse, Bp: Base pairs, PCR: Polymerase chain reaction, E. coli: Escherichia coli, EHEC: Enterohemorrhagic Escherichia coli, EPEC: Enteropathogenic Escherichia coli, ETEC: Enterotoxigenic Escherichia coli, EIEC: Enteroinvasive Escherichia coli, EAggEC: Enteroaggregative Escherichia coli, DAEC: Diffuse adherent Escherichia coli
The reaction mixture (PCR master mix), 25 µL, was prepared with OmniMix HS (contains 3U TaKaRa hot start Taq polymerase, 200 µM dNTP, 4 mM MgCl2, 25 mM HEPES buffer pH 8.0 ± 0.1), 2 pmol of each primer, and 5 µL of template DNA. The PCR process involved 35 cycles, where each cycle comprised 1.5 min at 94°C for denaturation, followed by 1.5 min at 60°C for primer annealing, and finally, 1.5 min at 72°C for strand elongation.
PCR products were observed through the utilization of 1.5% agarose gel electrophoresis combined with ethidium bromide staining for visualization, and the gel pictures taken. A 100 base pair (bp) size ladder (1500–100 bp) was used to identify PCR amplicons. The positive controls used were 011: 011 (stx1 and stx2), 2348/69 (bfp and eae), H10407 (st and lt), EI-34 Nal-R (ipaH and virF), EI 34 Strep-R (ipaH and virF), O42 (aafII), and F-1845 (daaE) while S. flexineri and PCR grade water were used as negative controls.
Statistical analysis
Data were analyzed using the Z-test involving the use of the unweighted-pair group method. The statistical significance threshold was established at P ≤ 0.05 also in accessing the manifestation of toxin genes by the various strains of E. coli.
RESULTS
Detection of E. coli pathotypes and their virulence toxins using real-time and conventional PCR
The real-time and conventional PCR analysis of the 204 E. coli strains from the diarrheal stool and water expressed different compositions of toxins: Stx2, stx1, eae, bfp, st11, lt, virF, aafII, daaE, and uidA with an exception of ipaH toxin, as also observed from the positive standard [Figure 2], the gene for EIEC, which was not detected [Figure 3]. A total of 143 toxins (101 from fecal and 42 from water samples) and uidA were detected from the screened E. coli strains. The six categories of diarrheagenic E. coli with their corresponding toxins (enterohemorrhagic (stx1, stx2), enteropathogenic (eae, bfp), enterotoxigenic (stII and lt), enteroinvasive (virF and), enteroaggregative (aafII), and diffuse adherent including the less common DAEC with its corresponding toxin (daaE)) were identified [Table 2]. EAggEC toxin, aafII 2/143 (1.39%), EIEC toxin virF 2/143 (1.39%), and DAEC toxin daaE 2/143 (1.39%) were the least toxins detected as shown in Table 2. AafII 2/143 (1.39%) and virF 2/143 (1.39%) toxins were detected only from water isolates. UidA toxins were detected in all (stool and water) isolates, [Figure 1] followed by EPEC toxin, 85/143 (59.44%) of which 68 (33.33%) were from stool strains while 17 (8.33%) were from water strains [Figure 3]. While bfp was found only in stool E. coli strains, eae, lt, and st11 were found more in stool strains. More of the Shiga toxins (stx2) were isolated from the water strains 9/143 (21.42%) than the stool strains 2/143 (1.98%). Out of the 143 (4.90%) detected toxins, 7 were Stx1 of which 2/143(1.98%) were detected in stool strains while 5 (11.90%) were detected in water strains. The percentage of virulence toxins detected from all the samples (stool and water) are in the following decreasing order: UidA 204/204 (100%) > eae 85/143 (59.44%) > lt 20/143 (13.99%) > stx2 11/143 (7.69%) > bfp 10/143 (4.49%%) > stx1 7/143 (4.90%) > st11 4/143 (2.79%) > virf 2/143 (1.39%) and aaf11 2/143 (1.39%), and daaE 2/143 (1.39%) as shown in Figure 4. All the toxins under investigation except ipaH were detected from water strains while virF, aaf11, and ipaH were not detected from the stool strains [Table 2]. The toxins detected from fecal samples are in the decreasing order of uidA 204/204 (100%) > eae 68/101 (67.33%) > lt 16/101 (15.84%) > bfp 9/101 (8.91%) > st11 3/101 (2.97%) > stx2 2/101 (1.98%) and stx1 2/101 (1.98%) > daaE 1/101 (0.99%) while the toxins detected from water samples are in the decreasing order of uidA (100%) > eae 17/42 (40.48%) > stx2 9/42 (21.42%) > stx1 5/42 (11.9%) > lt 4/42 (9.52%) > VirF 2/42 (4.76%) and aaF11 2/42 (4.76%), bfp 1/42 (2.38%), st11 1/42 (2.38%), daaE 1/42 (2.38%) [Figure 5]. The virulence toxins were detected most from enteropathogenic strains 95/143 (66.43%), followed by enterotoxigenic strains 24/143 (16.78%) and enterohemorrhagic strains 18/143 (12.59%) as evident in Table 2. The EIEC strain with virF, 2/143 (1.39%); EAggEC strains with aaf11, 2/143 (1.39%); and DAEC strains with daaE, 2/143 (1.39%) had the same percentage degree of occurrence [Figure 4].
| E. coli pathotype | Toxins expressed | % Number of each toxin expressed+uidA |
|---|---|---|
| EHEC | st×1 | 7 (4.90) |
| st×2 | 11 (7.69) | |
| EPEC | Eae | 85 (59.44) |
| Bfp | 10 (6.99) | |
| ETEC | Lt | 20 (13.99) |
| st11 | 4 (2.79) | |
| EIEC | virF | 2 (1.39) |
| ipaH | 0 | |
| EAggEC | aaf11 | 2 (1.39) |
| DAEC | daaE | 2 (1.39) |
| Total=6 | 10+uidA | 143+uidA |
E. coli: Escherichia coli, EHEC: Enterohemorrhagic Escherichia coli, EPEC: Enteropathogenic Escherichia coli, ETEC: Enterotoxigenic Escherichia coli, EIEC: Enteroinvasive Escherichia coli, EAggEC: Enteroaggregative Escherichia coli, DAEC: Diffuse adherent Escherichia coli
![Multiplex PCR gel analysis of the control [positive (reference strains) and negative strains]. The positive strains: 011 (stx1 and stx2), 2348/69 (bfp and eae), H10407 (st and lt), EI-34 Nal-R (ipaH and virF), EI 34 Strep-R (ipaH and virF); O42 (aafII) and F-1845 (daaE). Lanes 1 and 10, 100bp sized ladder; lane 2, negative control (PCR grade water); lane 3, 042; lane 4, EI-34 Nal-R; lane 5, EI 34 Strep-R; lane 6, 933J; lane 7, F-1845; lane 8, 2348/69 and lane 9, H10407.](/content/127/2025/4/1/img/AJPPS-4-3-g002.png)
- Multiplex PCR gel analysis of the control [positive (reference strains) and negative strains]. The positive strains: 011 (stx1 and stx2), 2348/69 (bfp and eae), H10407 (st and lt), EI-34 Nal-R (ipaH and virF), EI 34 Strep-R (ipaH and virF); O42 (aafII) and F-1845 (daaE). Lanes 1 and 10, 100bp sized ladder; lane 2, negative control (PCR grade water); lane 3, 042; lane 4, EI-34 Nal-R; lane 5, EI 34 Strep-R; lane 6, 933J; lane 7, F-1845; lane 8, 2348/69 and lane 9, H10407.
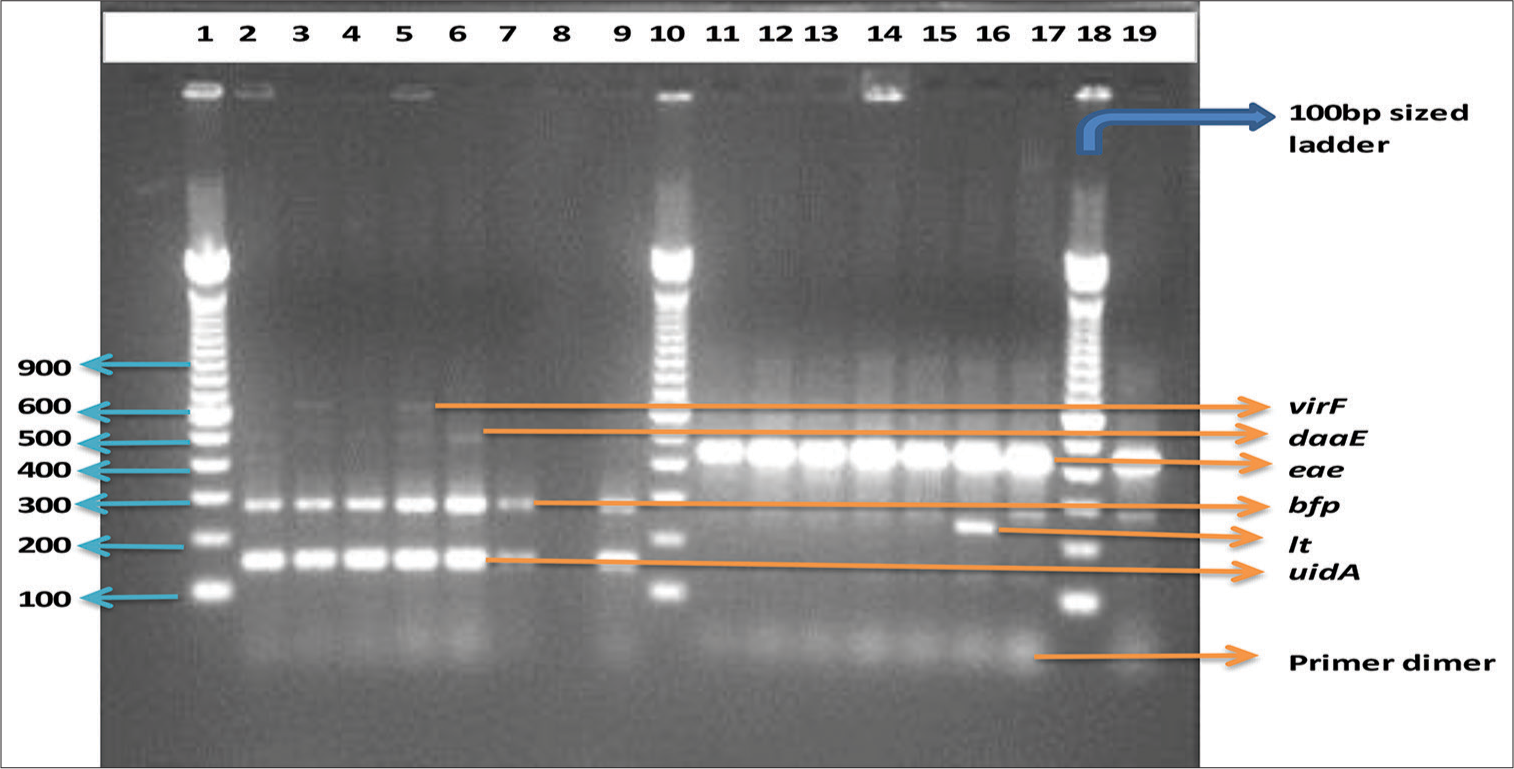
- Multiplex PCR gel analysis of diarrhoeagenic E. coli strains from Lagos and some selected reference strains. Lanes 1, 10 and 18 are 100-bp sized ladder; lane 19, bfp and eae positive control while lane 8 is the negative control (PCR grade water)
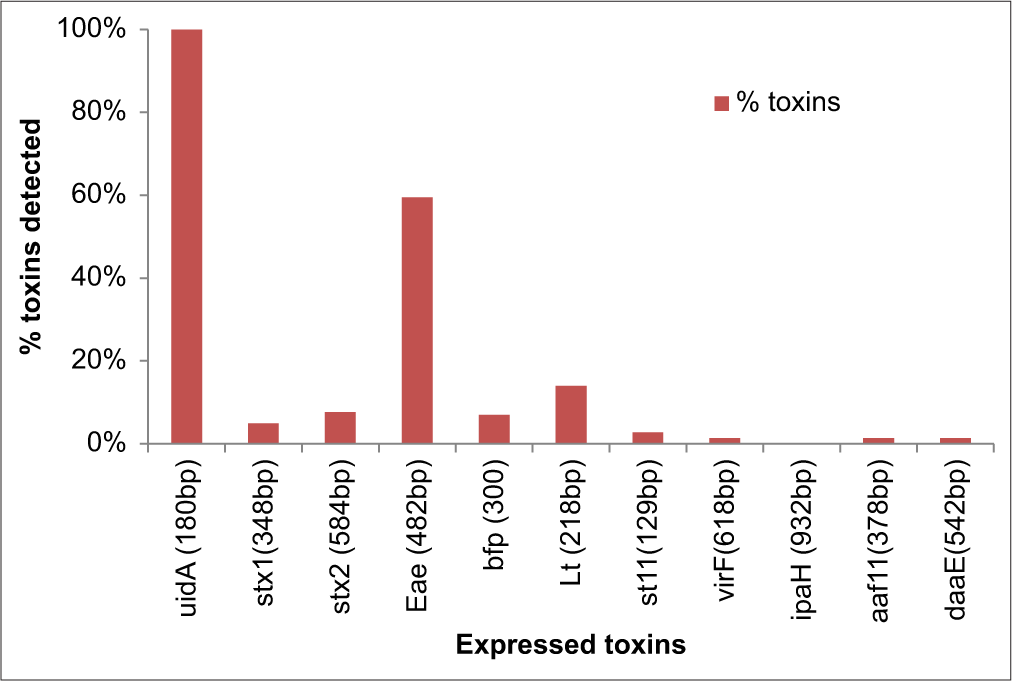
- Percentage Toxins detected from Escherichia coli isolated from all (Stool and water) samples.
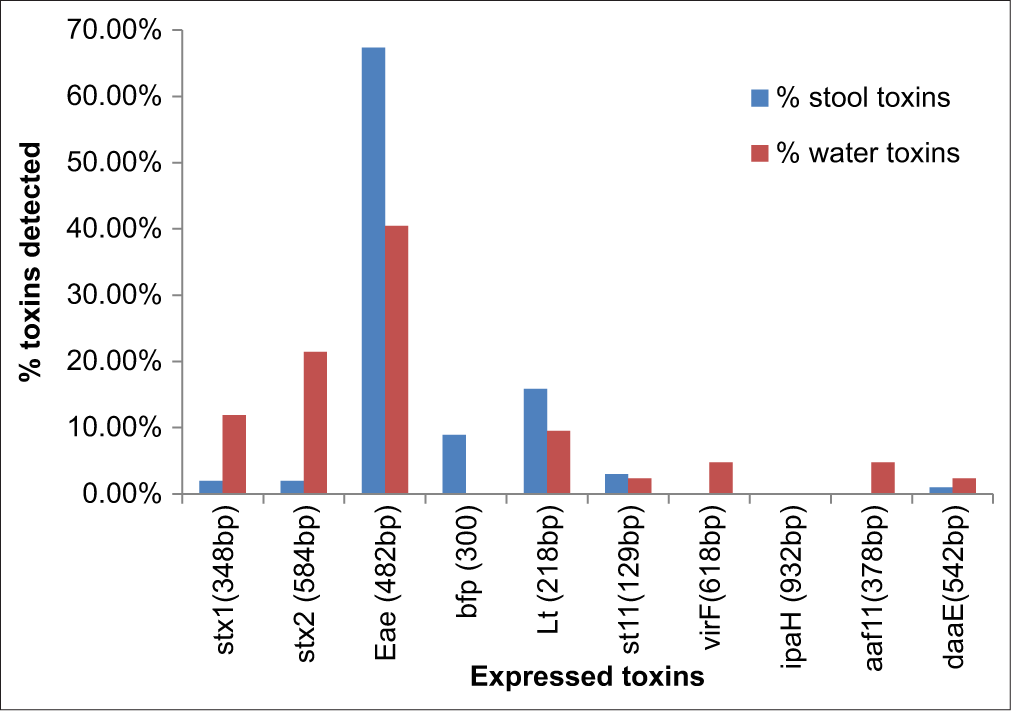
- Percentage Toxins detected from Escherichia coli isolated from water and diarrhoeal stool. uidA was 100% detected in both stool and water strains.
DISCUSSION
A broad spectrum of bacterial, viral, and parasitic pathogens can colonize the intestine irrespective of the presence or absence of pronounced dehydrating diarrhea.[11] The death burden associated with diarrhea can be comprehended in terms of the systematic malnutrition resulting from persistent diarrheal occurrences and chronic enteropathy induced by recurrent gastrointestinal infections. This phenomenon is conspicuous within developing nations, where the prevalence of diarrheal infections correlates with substandard sanitary infrastructure, notably reflected in the hygiene standards of ingested food and water sources, frequently compromised by diverse intestinal pathogens.[11] UidA was detected in all the E. coli strains, and its presence being an indicator toxin further confirms the studied strains as E. coli.[40,41] The detection of E. coli which is an indicator organism and its survival in any medium suggests contamination with other dangerous organisms such as Salmonella, Shigella, Vibrio species, Campylobacter species, Clostridium welchii, and Entamoeba histolytica enterovirus. Detection of the six diarrheagenic E. coli pathotypes (enterohemorrhagic, enteropathogenic, enterotoxigenic, enteroinvasive, enteroaggregative, and diffuse adherent) and their virulence toxins (uidA [100%], eae [42%], lt [9.8%], stx2 [5.4%], bfp [5%], stx1 [3.4%], st11 [2%], Virf [1%], Aaf11 [1%], and daaE [1%]) from the stool and water strains is a clear indication that more pathogenic organisms could be common in the environment. This poses a serious threat to people living in Lagos and to public health at large. Detection of 10 toxins (uidA [100%], eae 17/117 [14.52%], stx2 9/117 [7.69%], lt 4/117 [3.42%], stx1 5/117 [4.27%], VirF 2/117 [1.71%], aaF11 2/117 [1.71%], bfp [0.85%], st11 1/117 [0.85%], daaE 1/117 [0.85%]) out of 11 targeted E. coli toxins from water strains against the detected eight toxins (uidA [100%], eae 68/87 [78.16%], lt 16/87 [18.39%], bfp 9/87 [10.36%], stx2 2/87 [2.29%], st11 3/87 [3.45%], stx1 2/87 [2.29%], and daaE 1/87 [1.15%]) from the stool strains also exposes the degree of contamination in the studied environment. More of the stx2 toxins were expressed by the water strains 9/143 (6.29%) than in stool strains 2/142 (1.39%), a call for urgent attention to the need for various disinfection/inspection techniques and provision of potable water by government authorities and concerned agencies since only about 4 million of the 15 million population in Lagos have access to piped water.[42,43] It was observed that the toxins eae, lt, and bfp were significantly detected in stool (clinical) strains than the environmental (water) strains, indicating the strains were not just commensals but pathogenic which might have enhanced the disease burden of the patients. The prevalence of the eae toxin gene, responsible for encoding intimin, a pivotal intestinal adherence factor, exhibited extensive distribution across both clinical and environmental specimens. The identification of 8 diarrheagenic E. coli virulence toxin genes (uidA, eae, lt, bfp, stx2, stx1, ST11, and daaE) in the diarrheal stool samples and 10 virulence toxin genes (uidA, eae, stx2, lt, stx1, VirF, aaF11, bfp, st11, daaE) from water isolates [Figure 3] is of great interest. It is scientifically sound to postulate that in the context of potable water procurement, the application of relative filtration techniques may occur without substantial alteration to bacterial concentrations within the water source.
The city of Lagos exhibits compelling indications of the widespread and indiscriminate utilization of antibiotics.[44] which is a major factor that predisposes antibiotic resistance.[45,46] The discovery of extensive toxin gene expression by E. coli isolates in both environmental and fecal samples raises significant public health concerns. These findings indicate an augmented burden on patients suffering from the affliction of virulence toxin genes. There is a great necessity for understanding the prevalence and expression patterns of these genes for establishing targeted interventions to mitigate the health risks associated with these pathogens. Great significance is attached to this result because of the prominent position of diarrheagenic E. coli among bacterial pathogens contributing to diarrheal illness.
CONCLUSION
The conventional and real-time PCR studies of E. coli strains from Lagos, Nigeria, detected uidA as an inherent gene in all the strains of E. coli. Enteroinvasive (VirF and ipaH) E. coli pathotype is not commonly detected but virF was detected in environmental strain, an indication of an increased unhygienic environment. The environmental E. coli strains exhibited more toxins than the stool strains. Shiga toxins (Stx1 and Stx2) were isolated more from the environmental samples compared to the stool samples which are indications of the accessible water samples in Lagos being a threat to human health. The most predominant pathotypes detected with their expressed toxins were EPEC and ETEC.
Ethical approval
The research/study was approved by the Institutional Review Board at the Research Grants and Experimentation Ethics Committee, number CM/COM/08/VOL.XXV, dated February 20, 2014.
Declaration of patient consent
The authors certify that they have obtained all appropriate patient consent.
Conflicts of interest
There are no conflicts of interest.
Use of artificial intelligence (AI)-assisted technology for manuscript preparation
The authors confirm that there was no use of artificial intelligence (AI)-assisted technology for assisting in the writing or editing of the manuscript and no images were manipulated using AI.
Financial support and sponsorship
None.
References
- Comparison of multiplex PCR with serogrouping and PCR-RFLP of fliC gene for the detection of enteropathogenic Escherichia coli (EPEC) Braz J Infect Dis. 2011;15:365-369. doi: 10.1016/S1413-8670(11)70206-9
- [CrossRef] [PubMed] [Google Scholar]
- The global problem of childhood diarrhoeal diseases: Emerging strategies in prevention and management. Ther Adv Infect Dis. 2018;5:29-43. doi: 10.1177/2049936117744429
- [CrossRef] [PubMed] [Google Scholar]
- Childhood diarrhoeal diseases in developing countries. Heliyon. 2020;6:e03690. doi: 10.1016/j.heliyon.2020.e03690
- [CrossRef] [PubMed] [Google Scholar]
- Water, sanitation and hygiene. 2021. Available from: https://www.unicef.org/wca/what-we-do/wash [Last accessed on 2021 Nov 23]
- [Google Scholar]
- Aetiology of diarrhoea and virulence properties of diarrhoeagenic Escherichia coli among patients and healthy subjects in southeast Nigeria. J Health Popul Nutr. 2010;28:245-252. doi: 10.3329/jhpn.v28i3.5551
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of diarrheal diseases and associated factors among under-five children in Dale District, Sidama zone, Southern Ethiopia: a cross-sectional study. BMC Public Health. 2019;19:1235. doi: 10.1186/s12889-019-7579-2
- [CrossRef] [PubMed] [Google Scholar]
- Etiology of acute diarrhea in adults in Southwestern Nigeria. J Clin Microbiol. 2003;41:4525-4530. doi: 10.1128/JCM.41.10.4525-4530.2003
- [CrossRef] [PubMed] [Google Scholar]
- Antibiotic resistance patterns of intestinal Escherichia coli isolates from Nicaraguan children. J Med Microbiol. 2011;60:216-222. doi: 10.1099/jmm.0.020842-0
- [CrossRef] [PubMed] [Google Scholar]
- Diarrheagenic Escherichia coli in acute gastroenteritis in infants in North-West Italy. New Microbiol. 2011;34(1):45-51.
- [Google Scholar]
- Typical enteroaggregative Escherichia coli is the most prevalent pathotype among E. coli strains causing diarrhea in Mongolian children. J Clin Microbiol. 2004;42:133-139. doi: 10.1128/JCM.42.1.133-139.2004
- [CrossRef] [PubMed] [Google Scholar]
- Preponderance of toxigenic Escherichia coli in stool pathogens correlates with toxin detection in accessible drinking-water sources. Epidemiol Infect. 2015;143:494-504. doi: 10.1017/S0950268814001046
- [CrossRef] [PubMed] [Google Scholar]
- Global mortality associated with 33 bacterial pathogens in 2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet Lond Engl. 2022;400:2221-2248. doi: 10.1016/S0140-6736(22)02185-7
- [CrossRef] [Google Scholar]
- Prevalence of diarrheal disease and associated factors among under-five children in flood-prone settlements of Northwest Ethiopia: A cross-sectional community-based study. Front Pediatr. 2023;11:1056129. doi: 10.3389/fped.2023.1056129
- [CrossRef] [PubMed] [Google Scholar]
- The rise of diarrheal illnesses in the children of Pakistan amidst COVID-19: A narrative review. Health Sci Rep. 2023;6:e1043. doi: 10.1002/hsr2.1043
- [CrossRef] [PubMed] [Google Scholar]
- Enteric pathogens risk factors associated with household drinking water: A case study in Ugu District Kwa-Zulu Natal Province, South Africa. Int J Environ Res Public Health. 2022;19:4431. doi: 10.3390/ijerph19084431
- [CrossRef] [PubMed] [Google Scholar]
- Association between microbial water quality, sanitation and hygiene practices and childhood diarrhea in Kersa and Omo Nada districts of Jimma Zone, Ethiopia. PLoS One. 2020;15:e0229303. doi: 10.1371/journal.pone.0229303
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence and factors associated with diarrheal diseases among children below five years in selected slum settlements in Entebbe municipality, Wakiso district, Uganda. BMC Pediatr. 2022;22:394. doi: 10.1186/s12887-022-03448-2
- [CrossRef] [PubMed] [Google Scholar]
- Diarrhoeal disease. Available from: https://www.who.int/news-room/fact-sheets/detail/diarrhoeal-disease [Last accessed on 2024 Jun 05]
- [Google Scholar]
- Focus on acute diarrhoeal disease. World J Gastroenterol. 2009;15:3341-3348. doi: 10.3748/wjg.15.3341
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and self-treatment of travelers’ diarrhea in a large, prospective cohort of department of defense beneficiaries. J Travel Med. 2015;22:152-160. doi: 10.1111/jtm.12179
- [CrossRef] [PubMed] [Google Scholar]
- Epidemiology and etiology of Traveler’s diarrhea in Bangkok, Thailand, a case-control study. Trop Dis Travel Med Vaccines. 2019;5:9. doi: 10.1186/s40794-019-0085-9
- [CrossRef] [PubMed] [Google Scholar]
- Escherichia coli as commensal and pathogenic bacteria among food-producing animals: Health implications of extended spectrum β-lactamase (ESBL) production. Animals. 2020;10:2239. doi: 10.3390/ani10122239
- [CrossRef] [PubMed] [Google Scholar]
- Single multiplex PCR assay to identify simultaneously the six categories of diarrheagenic Escherichia coli associated with enteric infections. J Clin Microbiol. 2005;43:5362-5365. doi: 10.1128/JCM.43.10.5362-5365.2005
- [CrossRef] [PubMed] [Google Scholar]
- Identification of diarrheagenic Escherichia coli isolated from infants and children in Dar es Salaam, Tanzania. BMC Infect Dis. 2007;7:92. doi: 10.1186/1471-2334-7-92
- [CrossRef] [PubMed] [Google Scholar]
- Modulation of the enterohemorrhagic E. coli virulence program through the human gastrointestinal tract. Virulence. 2013;4:315-323. doi: 10.4161/viru.24318
- [CrossRef] [PubMed] [Google Scholar]
- Enteropathogenic Escherichia coli infection in children. Curr Opin Infect Dis. 2011;24:478-483. doi: 10.1097/QCO.0b013e32834a8b8b
- [CrossRef] [PubMed] [Google Scholar]
- The intriguing evolutionary journey of enteroinvasive E. coli (EIEC) toward pathogenicity. Front Microbiol. 2017;8:2390. doi: 10.3389/fmicb.2017.02390
- [CrossRef] [PubMed] [Google Scholar]
- Low distribution of genes encoding virulence factors in Shigella flexneri serotypes 1b clinical isolates from eastern Chinese populations. Gut Pathog. 2017;9:76. doi: 10.1186/s13099-017-0222-9
- [CrossRef] [PubMed] [Google Scholar]
- Identification and characterisation of enteroaggregative Escherichia coli subtypes associated with human disease. Sci Rep. 2020;10:7475. doi: 10.1038/s41598-020-64424-3
- [CrossRef] [PubMed] [Google Scholar]
- Enteroaggregative Escherichia coli pathotype: A genetically heterogeneous emerging foodborne enteropathogen. FEMS Immunol Med Microbiol. 2012;66:281-298. doi: 10.1111/j.1574-695X.2012.01008.x
- [CrossRef] [PubMed] [Google Scholar]
- Genotyping of enteroaggregative Escherichia coli and identification of target genes for the detection of both typical and atypical strains. Diagn Microbiol Infect Dis. 2006;55:13-19. doi: 10.1016/j.diagmicrobio.2005.10.019
- [CrossRef] [PubMed] [Google Scholar]
- Enteroaggregative Escherichia coli (EAEC): A cause of acute and persistent diarrhea of worldwide importance. J Infect Dis. 2010;202:503-505. doi: 10.1086/654895
- [CrossRef] [PubMed] [Google Scholar]
- Diffusely adherent Escherichia coli strains isolated from children and adults constitute two different populations. BMC Microbiol. 2013;13:22. doi: 10.1186/1471-2180-13-22
- [CrossRef] [PubMed] [Google Scholar]
- Performance standards for antimicrobial susceptibility testing United States: CLSI; 2017.
- [Google Scholar]
- Enumeration of Escherichia coli and the Coliform Bacteria In: FDA's bacteriological analytical manual, Ch. 4. FDA. 2020. Available from: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-4-enumeration-escherichia-coli-and-coliform-bacteria [Last accessed on 2024 Jun 12]
- [Google Scholar]
- Molecular serotyping and antibiotic resistance patterns of Escherichia coli isolated in hospital catering service in Morocco. Int J Microbiol. 2020;2020:5961521. doi: 10.1155/2020/5961521
- [CrossRef] [PubMed] [Google Scholar]
- Prevalence of nonO157:H7 shiga toxin-producing Escherichia coli in diarrheal stool samples from Nebraska. Emerg Infect Dis. 2000;6:530-533. doi: 10.3201/eid0605.000513
- [CrossRef] [PubMed] [Google Scholar]
- Detection and characterization of verocytotoxin-producing Escherichia coli by automated 5' nuclease PCR assay. J Clin Microbiol. 2003;41:2884-2893. doi: 10.1128/JCM.41.7.2884-2893.2003
- [CrossRef] [PubMed] [Google Scholar]
- Molecular characterization of diarrheagenic Escherichia coli isolated from meat products sold at Mansoura city, Egypt. Food Control. 2012;25:159-164. doi: 10.1016/j.foodcont.2011.10.026
- [CrossRef] [Google Scholar]
- Distribution of uidA gene sequences in Escherichia coli isolates in water sources and comparison with the expression of beta-glucuronidase activity in 4-methylumbelliferyl-beta-D-glucuronide media. Appl Environ Microbiol. 1993;59:2271-2276. doi: 10.1128/aem.59.7.2271-2276.1993
- [CrossRef] [PubMed] [Google Scholar]
- Species specific PCR based detection of Escherichia coli from Indian foods. 3 Biotech. 2017;7:130. doi: 10.1007/s13205-017-0784-8
- [CrossRef] [PubMed] [Google Scholar]
- The transition to a predominantly urban world and its underpinnings. (Human settlements discussion paper series).
- [Google Scholar]
- Urbanization, cities, and health: The challenges to Nigeria-a review. Ann Afr Med. 2017;16:149-158. doi: 10.4103/aam.aam_1_17
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial resistance awareness and antibiotic prescribing behavior among healthcare workers in Nigeria: A national survey. BMC Infect Dis. 2021;21:22. doi: 10.1186/s12879-020-05689-x
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial resistance: A global multifaceted phenomenon. Pathog Glob Health. 2015;109:309-318. doi: 10.1179/2047773215Y.0000000030
- [CrossRef] [PubMed] [Google Scholar]
- Antimicrobial resistance: A growing serious threat for global public health. Healthcare. 2023;11:1946. doi: 10.3390/healthcare11131946
- [CrossRef] [PubMed] [Google Scholar]







